News & Highlights
Topics: Bioinformatics, In the News, Open-source Data Tools, Technology
Searching COVID-19 Publications
International website links researchers, collaborators.
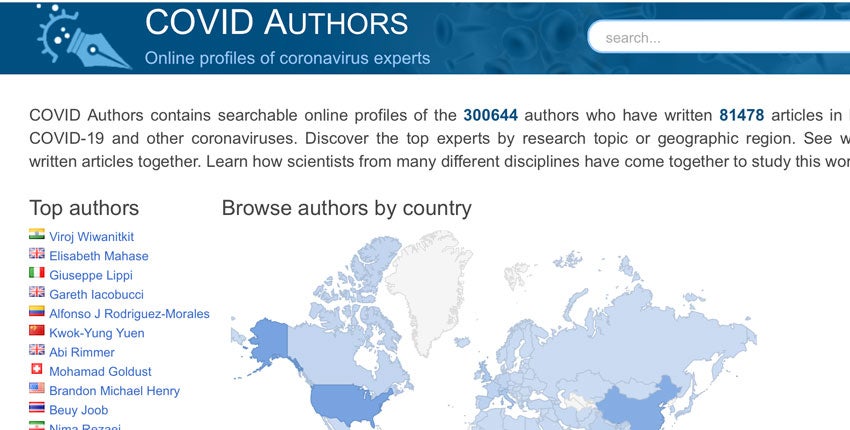
Faced by the growing enormity of the coronavirus pandemic, the global research community wasted no time. By the end of December 2020, more than 300,000 researchers from different countries and fields of inquiry have authored more than 81,000 publications on SARS-CoV-2 and COVID-19.
Now, the profiles of all these coronavirus experts and their network of collaborators are easily searchable through a newly released platform called COVIDAuthors. The website is freely accessible and is geared specifically to the needs of researchers looking for collaborators and for policymakers seeking local experts to consult.
“At the beginning of the pandemic, it was obvious to the scientific community that we needed to come together to solve this complicated, global problem,” said the creator of COVIDAuthors, Griffin Weber, an associate professor of medicine and of bioinformatics at Harvard Medical School (HMS) and Beth Israel Deaconess Medical Center.
“At the beginning of the pandemic, it was obvious to the scientific community that we needed to come together to solve this complicated, global problem.”
COVIDAuthors is powered by the same software behind Harvard Catalyst Profiles, a site which features profiles and publications of faculty from HMS, Harvard School of Dental Medicine, and the Harvard T.H. Chan School of Public Health.
With a background in medical informatics, Weber is involved in several studies that are leveraging hospitals’ clinical databases to learn more about COVID-19’s disease course.
“We’re able to tap into databases around the world, but to increase the number of patients, we needed to find additional collaborators and hospitals to participate,” Weber said. “We realized that our Profiles software could have a role in facilitating this process.”
The COVIDAuthors website downloads the National Library of Medicine’s LitCOVID abstracts and all PubMed articles about the virus and COVID-19. The Profiles software then automatically uses computer algorithms to find and pull together other data from various sources to create a profile page for each author.
“It’s all about connections between people and visualizing those connections,” Weber explained, saying it goes beyond listing backgrounds and affiliations.
A results sidebar shows the top research areas people work in, other members on their teams, what departments they work in, and other such connections.
Profiles was created as one of Harvard Catalyst’s goals when it was launched in 2008, to bring together the Harvard community, which had been siloed by hospitals or disciplines, explained Weber.
“One thing that is very necessary for a community is to find each other,” he said. “So we built the Profiles website in 2008 with about 24,000 faculty from the medical school, dental school, and school of public health.”
In addition to helping researchers find collaborators, he said, Profiles has been used by students to find mentors, administrators to form committees, course directors to find other teachers, and patients to find clinicians.
“It’s the most widely used tool to come out of Harvard Catalyst,” he said. In the past year, the site has been visited more than 3.1 million times.
“It’s all about connections between people and visualizing those connections.”
Within Harvard Catalyst, he pointed out, “We generally develop first for Harvard, which, because of our multiple academic medical centers and hospitals, provides a model for scaling up to the rest of the country.”
Profiles is freely available to other institutions that want to adopt it. Dozens of other organizations now use the open-source program, have uploaded their own faculty to it, and have contributed their own coding and functionality.
“So it’s really a community-built tool,” Weber said.
Weber also created the original prototype software for Harvard’s Shared Health Research Information Network (SHRINE), which connects de-identified patient databases across multiple institutions.
This year, because of his expertise in hospital clinical databases, Weber was also part of a large international consortium with more than 100 investigators and 350 hospitals called the 4CE, which is doing studies to characterize COVID-19 using electronic health records. Because of his involvement in this study, Weber has a profile on COVIDAuthors, too.
“Like everyone else, I’d never researched this virus before, but I have an expertise that is relevant and joined a scientific team that has generated new results related to the pandemic,” he said. “This website shows all the different kinds of people who have some kind of expertise that may be useful to teams addressing the COVID-19 challenge.”
The website was created for SARS-CoV-2 and COVID-19 research, but sites using Profiles can also be created around any topic, he pointed out. Key to doing this, he said, is the author disambiguation engine, which sorts out whether an individual named on one paper, for example, is the same as an individual with the same name identified on another publication.
“We can imagine the same type of website could be used for other global pandemics or medical problems that affect people globally,” he said, “such as hypertension, diabetes, and cancer.”

